Identifying Shells using Convolutional Neural Networks: Data Collection and Model Selection
Published on: 29 December 2024
Abstract
This study explores the use of convolutional neural networks (CNNs) in shell identification, a critical task in taxonomy and biodiversity monitoring. By leveraging a vast dataset of over 1.6 million shell images representing more than 58,000 species, the research demonstrates the ability of CNNs to achieve high accuracy in classifying shell species. EfficientNetV2 models were evaluated, with EfficientNetV2 B2 selected for its balance between performance, training speed, and computational efficiency. However, the accuracy of the models is affected by the quality of the images and the number of images available for each species. Challenges such as dataset imbalance and model overfitting are addressed, highlighting the need for strategies like synthetic augmentation and multimodal approaches. The results underscore the potential of CNN-based tools to streamline species identification, support ecological research, shell collection, and empower citizen science initiatives, while identifying avenues for further refinement in data quality, model robustness, and computational efficiency.
Introduction
Deep learning, a subfield of machine learning, focuses on developing algorithms that emulate human cognitive processes for problem-solving. Deep learning techniques have gained prominence in various fields, including computer vision, where Convolutional Neural Networks (CNNs) have emerged as a powerful tool. CNNs excel in automatically learning and extracting complex features from images, leading to significant advancements in image classification, object detection, and image segmentation. Image classification is fundamental to organism identification, and CNNs have significantly improved the accuracy and efficiency of this task. In image classification, CNNs are trained to categorize images into predefined classes, such as different species of plants or animals.
CNNs have demonstrated remarkable potential in identifying various organisms, including bacteria, plants, fish, and insects. For instance, researchers have successfully employed CNNs to classify bacterial species with high accuracy. One study reported a CNN model achieving an impressive 99.35% accuracy in identifying 33 different bacterial species [1, 2, 3].
In plant science, CNNs have been applied to automatically identify crop diseases. These applications can serve as valuable tools for assisting experts or for automated screening, contributing to more sustainable agricultural practices and improved food production security [4]. CNNs have proven effective in insect sound recognition. By analyzing spectrograms, which are visual representations of sound frequencies, CNNs can accurately identify different insect species. This capability has applications in ecological monitoring, pest control, and biodiversity studies [5].
While research on CNN-based identification of mollusca is an emerging field, some studies have explored the use of CNNs for identifying cephalopod species based on their beaks. This model utilized pre-trained CNNs (VGG19, InceptionV3, and ResNet50) to extract deep features from beak images. The study found that deep features were more accurate than traditional morphometric features in highlighting differences between cephalopod species. [6]
Despite their successes, CNN models have limitations. One major limitation is the requirement for large amounts of labeled training data. Obtaining and labeling such datasets can be time-consuming and expensive. Another limitation is the potential for overfitting, where the model performs well on the training data but poorly on unseen data. This can occur when the model is too complex or when the training data is not representative of the real-world data. Additionally, CNN models can be sensitive to variations in image quality, lighting conditions, and viewpoint. These variations can affect the model's ability to accurately identify organisms [7].
Mollusks, a diverse and ecologically significant phylum, represent a challenging group for identification due to their vast species richness, morphological plasticity, and the frequent presence of cryptic species. Traditional methods of mollusk identification rely heavily on morphological characteristics, often requiring specialized taxonomic expertise and manual examination of specimens. These methods can be time-consuming, prone to subjectivity, and particularly challenging for closely related species exhibiting subtle morphological differences. This problem is a challenge for all shell collectors.
In this text we describe how a large shell image dataset was collected and processed to make ready for analysis by a CNN. At the time of writing images of more than 50 000 species are collected, with more 1 600 000 images in total.
Methods and Results
Data Collection
Shell images were collected from many online resources, from specialized websites on shell collecting to institutes and universities. One of the largest collections of shell images is available on GBIF. Also online marketplace such as ebay contain a large collection of images. Other large shell image collections are available at , Malacopics, Femorale and Thelsica. A shell dataset created for AI is available [8].
Some online resources have facilities to download images, but most websites require a specialized webscraper. Scrapy , an open source and collaborative framework for extracting the data from websites, is used to create a custom webscraper to extract images and their scientific names. All data was stored in a MySQL database before further processing was performed.
Preprocessing
All names were checked against WoRMS or MolluscaBase for their validity. Names that were not found in WoRMS/MolluscaBase were excluded for further processing. While a large part of this data quality step was automated, a manual verification (time-consuming) step was also included. In addition to text-based quality control, both automated and manual preprocessing steps were applied to the images. Shells were detected in all images and cut out of the original image, having only 1 shell on each image. Other objects on the raw images (labels, measures, hands holding a shell, etc.) were removed. When appropiate the background was changed to a uniform black background.
Dataset Composition
A total of 1 640 000 images were collected (status dec. 2024), representing 58 233 species. After elimination of all images where no valid scientific species name was found, 1 460 526 images are left for further processing. The dataset is unbalanced as shown in the chart below. Note that a logarithmic scale is used in the chart.
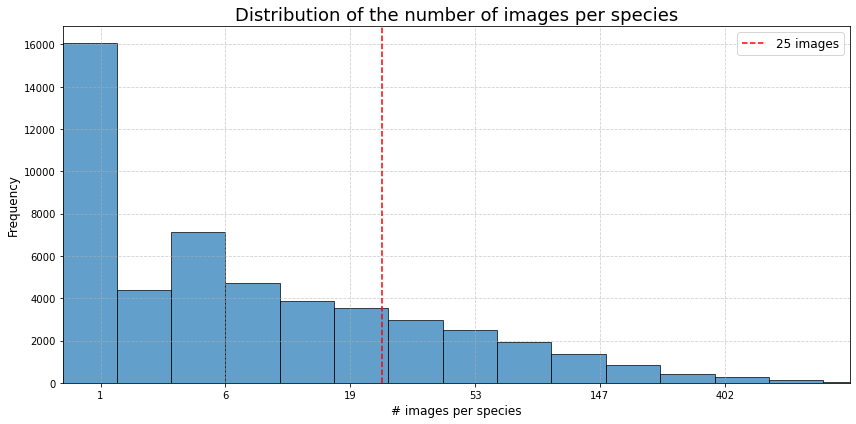
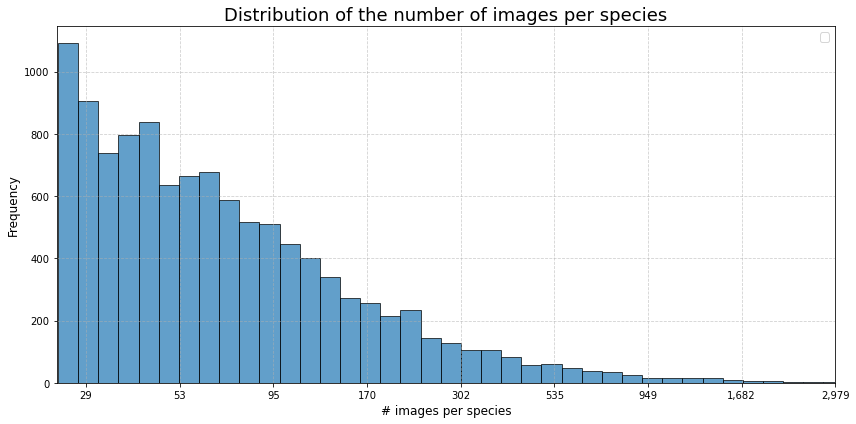
Number of images for each family (Top 15)
| Family | #images | #species with more than 25 images | Total #species |
|---|---|---|---|
| Cypraeidae | 157842 | 458 | 716 |
| Conidae | 119038 | 636 | 1165 |
| Muricidae | 62993 | 607 | 1722 |
| Olividae | 23290 | 149 | 349 |
| Pectinidae | 20934 | 163 | 367 |
| Nassariidae | 20899 | 245 | 624 |
| Volutidae | 20637 | 188 | 494 |
| Strombidae | 17857 | 90 | 163 |
| Mitridae | 16383 | 182 | 446 |
| Veneridae | 16204 | 190 | 546 |
| Trochidae | 15055 | 178 | 550 |
| Costellariidae | 14842 | 182 | 516 |
| Columbellidae | 12981 | 171 | 700 |
| Cardiidae | 12673 | 110 | 282 |
| Cymatiidae | 12176 | 88 | 130 |
Model selection
Recent research has shown that EfficientNetV2 models are a family of convolutional networks that achieve faster training speed and better parameter efficiency than previous models. This is achieved through a combination of training-aware neural architecture search and scaling, which jointly optimize training speed and parameter efficiency . This research also showed that EfficientNetV2 models can be trained efficiently at a large scale. They outperform Vision Transformers in terms of both accuracy and training efficiency. For example, by pretraining on the same ImageNet21k, EfficientNetV2 achieves 87.3% top-1 accuracy on ImageNet ILSVRC2012, outperforming the recent ViT by 2.0% accuracy while training 5x-11x faster using the same computing resources [9]. We are using Keras which has a large selection of CNN applications, including EfficientNetV2 models.
Accuracy and loss of the CNN models (Keras applications)
| Model | Size (Mb) | Training accuracy | Validation accuracy | Training loss | Validation loss | Epochs |
|---|---|---|---|---|---|---|
| EfficientNet B0 | 29 | 0.917 | 0.896 | 0.731 | 0.612 | 40 |
| EfficientNet B1 | 31 | 0.898 | 0.887 | 0.783 | 0.608 | 43 |
| EfficientNet B2 | 36 | 0.900 | 0.878 | 0.886 | 0.661 | 45 |
| EfficientNet B3 | 48 | 0.889 | 0.865 | 0.940 | 0.725 | 31 |
| EfficientNet B4 | 75 | 0.887 | 0.872 | 0.972 | 0.735 | 50 |
| EfficientNet B5 | 118 | 0.910 | 0.878 | 0.821 | 0.745 | 40 |
| EfficientNet B6 | 166 | 0.895 | 0.858 | 0.920 | 0.736 | 40 |
| EfficientNet B7 | 256 | 0.894 | 0.860 | 0.927 | 0.734 | 49 |
| EfficientNetV2 B0 | 29 | 0.893 | 0.882 | 0.927 | 0.639 | 32 |
| EfficientNetV2 B1 | 34 | 0.882 | 0.876 | 1.011 | 0.682 | 29 |
| EfficientNetV2 B2 | 42 | 0.873 | 0.888 | 1.052 | 0.670 | 41 |
| EfficientNetV2 B3 | 59 | 0.878 | 0.888 | 1.178 | 0.723 | 31 |
| EfficientNetV2 S | 88 | 0.858 | 0.865 | 1.178 | 0.749 | 48 |
| EfficientNetV2 M | 220 | 0.847 | 0.862 | 1.230 | 0.769 | 42 |
| EfficientNetV2 L | 479 | 0.830 | 0.841 | 1.458 | 0.881 | |
| All images of genus Harpa were used for to compare the models. The dataset contains 5837 images belonging to 13 classes (species). A 80:20 split was made for training and validation. The batch size is 64, learning rate is 0.0005. Class weight were given, early stopping was used. Three runs were made for each model, average taken. | ||||||
The results in the above table shows that the results between models are not significant different. A few EfficientNet models perform slightly worse, but the results do not help in the decision to select the optimal model to continue further training. Because performance metrics are similar, other factors like training time, computational resource requirements, and ease of deployment become crucial in deciding the best model. EfficientNet V2 models, for example, are designed to be more efficient and faster to train than their predecessors, which might be a deciding factor if performance differences are minimal. Among EfficientNet V2 models, a decision is made based on a balance between performance, training speed and model complexity. Considering these factors, EfficientNetV2 B2 was selected for further training and experimentation.
Class unbalance
Ideally, the dataset should contain an equal number of images for each class. However, in many seashell image datasets, this is not the case. Some classes may have significantly more images than others, leading to an unbalanced dataset. This imbalance can significantly impact the performance of the CNN model, causing it to be biased towards the majority class and perform poorly on the minority class.
Several techniques can be employed to address the challenges posed by unbalanced classes in image datasets including resampling, data augmentation,
class weighting, cost-senitive learning, transfer learning, ensemble methods and threshold moving. The keras implementation of EfficientNet includes transfer learning
with Imagenet. A model pre-trained on ImageNet can be fine-tuned on a smaller, imbalanced image dataset to improve its performance on the minority class.
Class weighting is also used when training the EfficienNet models which involves assigning different weights to different classes.
Higher weights are assigned to the minority class to give it more importance during the learning process. This can help to counter the bias towards
the majority class. The class weights are provided as a parameter when training the image dataset:
history = model.fit(train_data, epochs=epochs, validation_data=val_data, class_weight=classWeights, callbacks=[early_stopping, reduce_lr])When dealing with unbalanced classes, also a good choice of the hyperparameters is important. The batch size used during training can significantly impact the model's ability to learn from the minority class. Ideally, each batch should contain multiple minority class examples. The batch size should be several times greater than the imbalance ratio. For example, if the imbalance ratio is 10:1, then the batch size should be at least 50. A batch size of 64 was used in all training described in this article.
Discussion
The results indicate that CNNs can be used to identify shells with high accuracy. However, the accuracy of the models is affected by the quality of the images and the number of images available for each species. Future work will focus on improving the accuracy of the models by using more data and by developing more robust methods for handling image quality variations.
One of the primary obstacles encountered in this study was the inherent imbalance in the dataset. The uneven distribution of images among species reflects both ecological realities and biases in data availability, with common or visually distinctive species being overrepresented. While species with fewer than 25 images were excluded to ensure model reliability, this exclusion limits the applicability of the model to rarer species. Addressing this imbalance through targeted data collection or synthetic augmentation methods, such as generating images using generative adversarial networks (GANs), could enhance the comprehensiveness of the dataset.
The evaluation of EfficientNetV2 models illustrates trade-offs between accuracy, computational efficiency, and scalability. While the differences in performance metrics among the tested models were not substantial, the selection of EfficientNetV2 B2 for further experimentation was guided by practical considerations, such as training speed and resource demands. This decision underscores the importance of balancing technical and logistical factors in model deployment, particularly when working with large-scale datasets.
The relatively high training and validation accuracies achieved indicate that the model effectively learned to distinguish among shell species. However, the potential for overfitting remains a concern, especially given the controlled preprocessing steps that standardized image backgrounds and removed extraneous elements. Future work could explore the model’s robustness to variations in image quality, lighting, and orientation to better simulate real-world conditions.
Despite its successes, this study highlights several areas for improvement. The reliance on large, labeled datasets remains a bottleneck, as does the exclusion of species with limited image availability. Additionally, the focus on image-based identification does not account for other diagnostic features, such as location or ecological data, which could provide complementary insights. Future research could explore multimodal approaches combining image data with other types of information.
References
- [1] Farhana Sultana, A. Sufian, Paramartha Dutta. Advancements in Image Classification using Convolutional Neural Network. Fourth International Conference on Research in Computational Intelligence and Communication Networks (ICRCICN), 2018.
- [2] Ethan Yanjia Li 10 Papers You Should Read to Understand Image Classification in the Deep Learning Era. Towards Data Science, 2020
- [3] Shaily, Tumun & S., Kala. Bacterial Image Classification Using Convolutional Neural Networks. IEEE 17th India Council International Conference (INDICON), 2020
- [4] Boulent Justine et al. Convolutional Neural Networks for the Automatic Identification of Plant Diseases. Frontiers in Plant Science 10, 2019
- [5] Xue Dong et al. Insect Sound Recognition Based on Convolutional Neural Network. IEEE 3rd International Conference on Image, Vision and Computing (ICIVC), 2018
- [6] Hui Yuan Tan et al. Cephalopod species identification using integrated analysis of machine learning and deep learning approaches. PeerJ 9:e11825, 2021
- [7] Mai Ibraheam et al. A Performance Comparison and Enhancement of Animal Species Detection in Images with Various R-CNN Models. AI 2(4):552-577, 2021
- [8] Zhang, Q., Zhou, J., He, J. et al. A shell dataset, for shell features extraction and recognition.. Nature, Sci Data 6, 226 (2019)
- [9] Mingxing Tan and Quoc V. Le EfficientNetV2: Smaller Models and Faster Training. Proceedings of the 38th International Conference on Machine Learning, 2021.